Home > Research & Publications > EGFR Mutant Structural Database
EGFR Mutant Structural Database: computationally predicted 3D structures and the corresponding binding free energies with gefitinib and erlotinib
Abstract
Epidermal growth factor receptor (EGFR) mutation-induced drug resistance has caused great difficulties in the treatment of non-small-cell lung cancer (NSCLC). However, structural information is available for just a few EGFR mutants. In this study, we created an EGFR Mutant Structural Database (freely available at http://bcc.ee.cityu.edu.hk/data/EGFR.html), including the 3D EGFR mutant structures and their corresponding binding free energies with two commonly used inhibitors (gefitinib and erlotinib). We collected the information of 942 NSCLC patients belonging to 112 mutation types. These mutation types are divided into five groups (insertion, deletion, duplication, modification and substitution), and substitution accounts for 61.61% of the mutation types and 54.14% of all the patients. Among all the 942 patients, 388 cases experienced a mutation at residue site 858 with leucine replaced by arginine (L858R), making it the most common mutation type. Moreover, 36 (32.14%) mutation types occur at exon 19, and 419 (44.48%) patients carried a mutation at exon 21. In this study, we predicted the EGFR mutant structures using Rosetta with the collected mutation types. In addition, Amber was employed to refine the structures followed by calculating the binding free energies of mutant-drug complexes. The EGFR Mutant Structural Database provides resources of 3D structures and the binding affinity with inhibitors, which can be used by other researchers to study NSCLC further and by medical doctors as reference for NSCLC treatment.
Keywords: Epidermal growth factor receptor (EGFR), EGFR mutation database, Non-small-cell lung cancer (NSCLC), Tyrosine kinase inhibitor, Gefitinib, Erlotinib, Binding free energy
EGFR Mutant Structural Database
This epidermal growth factor receptor (EGFR) Mutant Structural Database contains 112 EGFR mutant types as well as the binding free energies with reversible tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib. Rosetta was employed to generate the 3D structures of the EGFR mutants based on the wild-type (WT) EGFR structures ("2ITY" and "1M17" from the Protein Data Bank). Amber was applied to optimize the mutant structures and compute the binding free energies with gefitinib and erlotinib.
Six PDB files (structureG01.pdb, structureG02.pdb, structureG03.pdb, structureE01.pdb, structureE02.pdb and structureE03.pdb) are available for each EGFR mutation type except L858R and G719S, for which the crystal structures can be downloaded from the Protein Data Bank. Structures G01, 02 and 03 were predicted based on WT EGFR "2ITY" while structures E01, 02 and 03 based on "1M17". We calculated the binding free energies of the structureG01-gefitinib complex and structureE01-erlotinib complex.
PDB files of all mutant structures can be downloaded here. Alternatively, the structure of each mutant can be found in the following table.
| Number | Mutation types | Description | 3D structures | Binding free energy with gefitinib (structureG01-gefitinib complex)(kcal/mol) | Binding free energy with erlotinib (structureE01-erlotinib complex)(kcal/mol) |
|---|---|---|---|---|---|
| A767_TLA_S768 | Insertion | PDB files | -42.3641 | -31.7393 | |
| D761_EAFQ_E762 | Insertion | PDB files | -31.0127 | -28.0779 | |
| D770_G_N771 | Insertion | PDB files | -23.9836 | -40.816 | |
| V769_ASV_D770 | Insertion | PDB files | -45.9188 | -33.6622 | |
| V769_CV_D770 | Insertion | PDB files | -36.1098 | -33.3974 | |
| V769_Y_D770 | Insertion | PDB files | -40.7622 | -33.868 | |
| V774_HV_C775 | Insertion | PDB files | -32.6008 | -38.1589 | |
| delE746_A750 | Deletion | PDB files | -35.2995 | -44.6007 | |
| delE746_S752 | Deletion | PDB files | -31.2015 | -33.2221 | |
| delE749_T751 | Deletion | PDB files | -42.381 | -38.8129 | |
| delL747_E749 | Deletion | PDB files | -31.3923 | -32.0382 | |
| delL747_S752 | Deletion | PDB files | -43.1211 | -35.4221 | |
| delL747_T751 | Deletion | PDB files | -28.4833 | -33.5532 | |
| dulA767_V769 | Duplication | PDB files | -34.318 | -34.6261 | |
| dulH773 | Duplication | PDB files | -41.753 | -36.7459 | |
| dulK739_I744 | Duplication | PDB files | -44.578 | -30.6392 | |
| dulN771_H773 | Duplication | PDB files | -42.6561 | -45.7011 | |
| dulP772_H773 | Duplication | PDB files | -45.2756 | -36.3212 | |
| dulS768_D770 | Duplication | PDB files | -37.428 | -33.2166 | |
| delE709_T710insD | Modification | PDB files | -48.128 | -38.1988 | |
| delE746_A750insAP | Modification | PDB files | -34.6648 | -37.0231 | |
| delE746_A750insQP | Modification | PDB files | -34.2928 | -25.7533 | |
| delE746_P753insLS | Modification | PDB files | -47.8951 | -37.3047 | |
| delE746_S752insA | Modification | PDB files | -43.8891 | -25.5718 | |
| delE746_S752insD | Modification | PDB files | -36.7967 | -40.6184 | |
| delE746_S752insV | Modification | PDB files | -42.7755 | -33.2645 | |
| delE746_T751insA | Modification | PDB files | -27.2134 | -41.3297 | |
| delE746_T751insI | Modification | PDB files | -42.2341 | -35.043 | |
| delE746_T751insV | Modification | PDB files | -34.0055 | -37.5001 | |
| delE746_T751insVA | Modification | PDB files | -34.5217 | -29.8218 | |
| delL747_A750insP | Modification | PDB files | -35.3201 | -39.7179 | |
| delL747_A755insSKG | Modification | PDB files | -29.3087 | -27.4069 | |
| delL747_K754insANKG | Modification | PDB files | -33.6135 | -33.9767 | |
| delL747_K754insSR | Modification | PDB files | -31.5194 | -39.2237 | |
| delL747_P753insQ | Modification | PDB files | -41.9879 | -38.0401 | |
| delL747_P753insS | Modification | PDB files | -28.5837 | -38.1157 | |
| delL747_S752insQ | Modification | PDB files | -36.6477 | -35.6391 | |
| delL747_T751insP | Modification | PDB files | -33.2542 | -39.8447 | |
| delL747_T751insPI | Modification | PDB files | -34.5882 | -33.3845 | |
| delL747_T751insQ | Modification | PDB files | -28.4329 | -34.3236 | |
| delL747_T751insS | Modification | PDB files | -44.2633 | -33.1996 | |
| delT751_I759insN | Modification | PDB files | -43.412 | -32.8137 | |
| delT751_I759insS | Modification | PDB files | -43.9421 | -33.0382 | |
| A763V | Substitution | PDB files | -39.2635 | -43.0264 | |
| A839T | Substitution | PDB files | -32.0284 | -41.0178 | |
| A859T | Substitution | PDB files | -42.4575 | -43.0478 | |
| A864T | Substitution | PDB files | -35.7166 | -40.2333 | |
| D761N | Substitution | PDB files | -22.2239 | -42.9582 | |
| E709A | Substitution | PDB files | -33.8316 | -39.9262 | |
| E709G | Substitution | PDB files | -34.7371 | -40.4049 | |
| E709K | Substitution | PDB files | -42.0845 | -45.3753 | |
| E709Q | Substitution | PDB files | -32.7873 | -43.4006 | |
| E709V | Substitution | PDB files | -30.9753 | -43.8952 | |
| E746K | Substitution | PDB files | -32.1456 | -43.9412 | |
| G719A | Substitution | PDB files | -34.6437 | -44.5366 | |
| G719C | Substitution | PDB files | -45.4477 | -43.724 | |
| G719R | Substitution | PDB files | -37.6231 | -37.7947 | |
| G719S | Substitution | PDB files | -35.4427 | -43.4964 | |
| G724S | Substitution | PDB files | -45.9446 | -46.3624 | |
| G735S | Substitution | PDB files | -31.6324 | -39.9289 | |
| G779F | Substitution | PDB files | -33.8423 | -42.2278 | |
| G810S | Substitution | PDB files | -31.6482 | -40.3096 | |
| G863D | Substitution | PDB files | -33.8733 | -41.5613 | |
| H773L | Substitution | PDB files | -36.8514 | -39.7405 | |
| H773R | Substitution | PDB files | -34.9159 | -40.9024 | |
| H835L | Substitution | PDB files | -42.5147 | -41.8518 | |
| H850N | Substitution | PDB files | -36.3505 | -44.7243 | |
| I715S | Substitution | PDB files | -43.4651 | -42.6452 | |
| I853T | Substitution | PDB files | -28.7007 | -41.4466 | |
| K757R | Substitution | PDB files | -40.3422 | -41.9819 | |
| K846R | Substitution | PDB files | -30.4882 | -43.5378 | |
| L703V | Substitution | PDB files | -36.4537 | -41.8784 | |
| L718P | Substitution | PDB files | -24.9425 | -41.7059 | |
| L730F | Substitution | PDB files | -40.2145 | -40.5323 | |
| L792P | Substitution | PDB files | -33.5893 | -42.4954 | |
| L798F | Substitution | PDB files | -30.1356 | -42.1674 | |
| L833V | Substitution | PDB files | -41.7871 | -39.4146 | |
| L838V | Substitution | PDB files | -32.5122 | -40.2593 | |
| L858M | Substitution | PDB files | -25.8424 | -43.8768 | |
| L858R | Substitution | PDB files | -46.0101 | -45.1344 | |
| L861Q | Substitution | PDB files | -32.2291 | -43.802 | |
| L861R | Substitution | PDB files | -41.6314 | -38.323 | |
| N700D | Substitution | PDB files | -31.2545 | -40.5408 | |
| N826S | Substitution | PDB files | -31.9999 | -40.3192 | |
| P733L | Substitution | PDB files | -29.6333 | -43.0825 | |
| P772R | Substitution | PDB files | -36.6468 | -41.9497 | |
| R776C | Substitution | PDB files | -44.7521 | -42.4406 | |
| R831H | Substitution | PDB files | -30.6892 | -43.5846 | |
| S720F | Substitution | PDB files | -37.2478 | -42.0389 | |
| S752Y | Substitution | PDB files | -30.5925 | -41.9819 | |
| S768I | Substitution | PDB files | -34.9202 | -40.8649 | |
| T751I | Substitution | PDB files | -28.0795 | -41.0402 | |
| T783A | Substitution | PDB files | -41.5648 | -42.7488 | |
| T790M | Substitution | PDB files | -36.4416 | -41.4281 | |
| T847I | Substitution | PDB files | -30.2238 | -43.8651 | |
| V742A | Substitution | PDB files | -41.2833 | -41.3265 | |
| V765A | Substitution | PDB files | -30.4874 | -42.2189 | |
| V769L | Substitution | PDB files | -29.4517 | -39.4975 | |
| V769M | Substitution | PDB files | -31.232 | -41.9498 | |
| V774M | Substitution | PDB files | -36.6284 | -43.3995 | |
| V851A | Substitution | PDB files | -31.9523 | -39.194 | |
| V851I | Substitution | PDB files | -28.9115 | -39.4173 | |
| E709A_G719A | Substitution | PDB files | -32.7568 | -44.8934 | |
| E709K_L858R | Substitution | PDB files | -34.5234 | -43.8563 | |
| G719A_L858R | Substitution | PDB files | -42.2028 | -41.3036 | |
| G719A_L861Q | Substitution | PDB files | -35.8614 | -45.7243 | |
| G719C_S768I | Substitution | PDB files | -44.4563 | -41.8073 | |
| G724S_L861Q | Substitution | PDB files | -35.6351 | -45.3965 | |
| L858R_L861F | Substitution | PDB files | -34.8422 | -40.7398 | |
| R776H_L858R | Substitution | PDB files | -33.2638 | -43.246 | |
| S768I_V774M | Substitution | PDB files | -43.5982 | -39.3976 | |
| T854A_L858R | Substitution | PDB files | -29.8618 | -36.6631 |
Procedure used to build the EGFR Mutant Structural Database
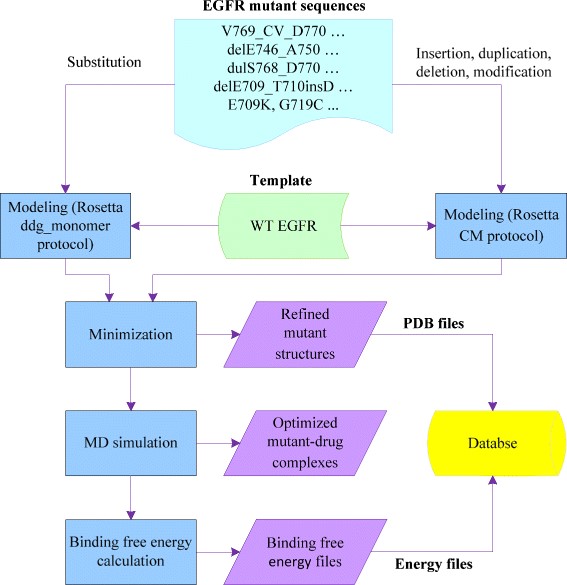
First, we applied Rosetta ddg_monomer protocol and comparative modeling (CM) protocol to predict EGFR mutant structures. Secondly, the predicted structures were refined with a minimization step using sander in Amber. Then a drug (gefitinib or erlotinib) was added to the mutant structures followed by MD simulation. Subsequently, we employed MM-GBSA in Amber to calculate the binding free energies of the EGFR mutants and the inhibitor. Finally, the refined mutant structures and their corresponding binding free energies with gefitinib and erlotinib were collected to establish the database.
[Back]